Quality of Data
According to Gartner: Poor Quality data weakens an organization’s competitive standing and business objectives
Data Wrangling Process
Process of transforming and mapping data from one raw data into another form with the intent of making it more appropriate and valuable for various task
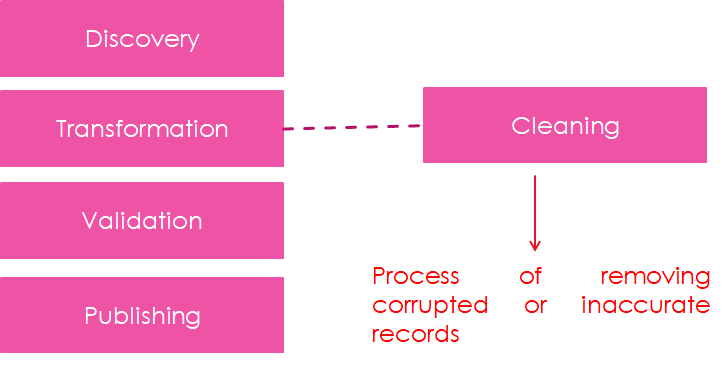
Data Cleaning Workflow
A typical data cleaning workflow includes:

Inspection
It includes,
- Detecting issues and errors
- Validating against rules and constraints
- Profiling data to inspect source data
- Visualizing data using statistical methods
Inspection
Data Profiling

Visualization
Visualizing the data using statistical methods can help to spot outliers
Cleaning
- Missing values can cause unexpected or biased results
- Duplicate data are data points that are repeated in your dataset
- Irrelevant data is data that is not contextual to the use case
- Data type conversion is needed to ensure that values in a field are stored as the data type of that field
- Syntax errors, such as white spaces, extra spaces , types and formats need to be fixed
- Outliers need to be examined for accuracy
Verification
It includes,
Inspecting results to establish effectiveness and accuracy achieved as a result of data cleaning
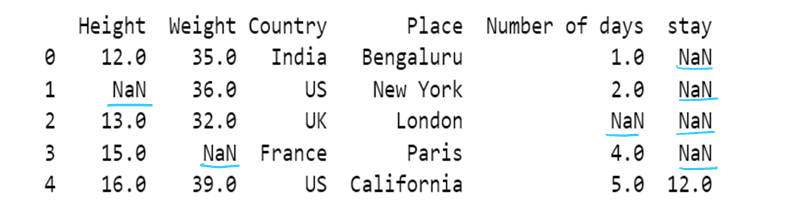
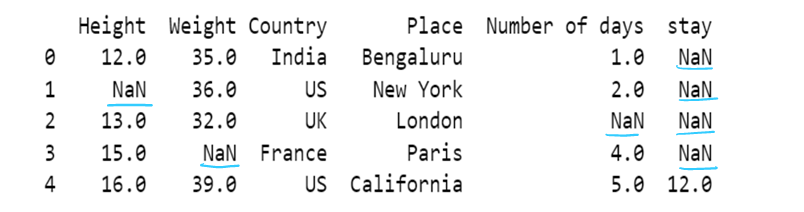
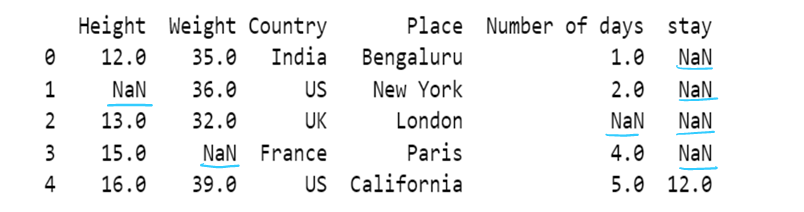
Handling Missing values
DataFrame.dropna(axis=0, how='any', thresh=None, subset=None, inplace=False)
Returns: DataFrame or None
DataFrame with NA entries dropped from it or None if inplace = Trueaxis : {0 or ‘index’, 1 or ‘columns’}, default 0Determine if rows or columns which contain missing values are removed.
0, or ‘index’ : Drop rows which contain missing values.
1, or ‘columns’ : Drop columns which contain missing value.
how:{‘any’, ‘all’}, default ‘any’
Determine if row or column is removed from DataFrame, when we have at least one NA or all NA.
‘any’ : If any NA values are present, drop that row or column.
‘all’ : If all values are NA, drop that row or column.
DataFrame.fillna(value=None, method=None, axis=None, inplace=False, limit=None, downcast=None)
Returns: DataFrame or None
Object with missing values filled or None if inplace = True
Value: scalar, dict, Series, or DataFrameValue to use to fill holes (e.g. 0), alternately a dict/Series/DataFrame of values specifying which value to use for each index (for a Series) or column (for a DataFrame). Values not in the dict/Series/DataFrame will not be filled. This value cannot be a list.
Method: {‘backfill’, ‘bfill’, ‘pad’, ‘ffill’, None}, default NoneMethod to use for filling holes in reindexed Series pad / ffill: propagate last valid observation forward to next valid backfill / bfill: use next valid observation to fill gap.
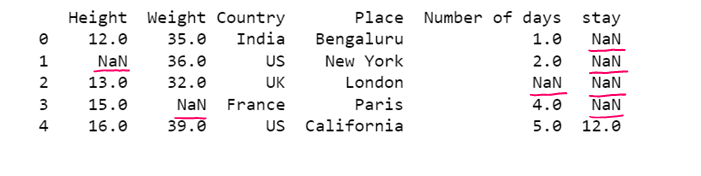
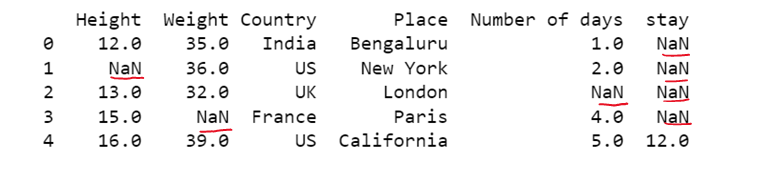
# Replacing all NaN with a given value
df1= dataset.fillna('E')
print(df1)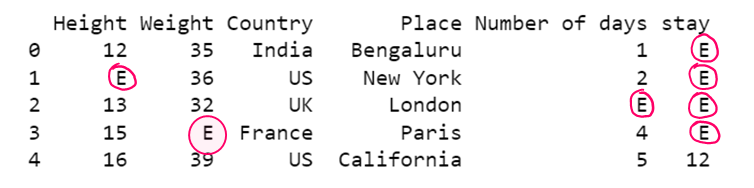
#b) Replacing NaN present in one specific column
dataset['stay'] = dataset['stay'].fillna(0)
dataset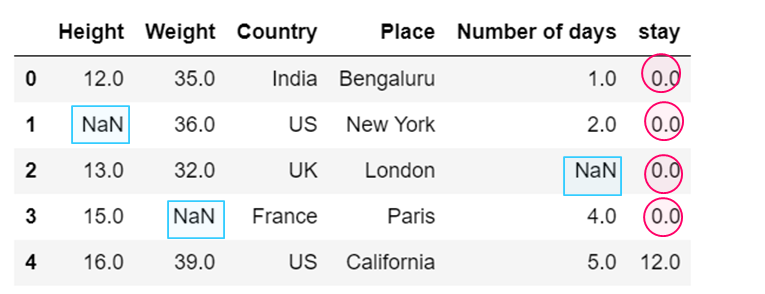
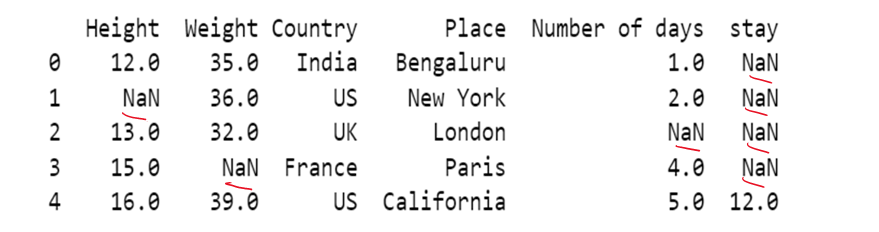
#b) using method attribute
df['Height']=df['Height'].fillna(method='backfill')
df['Weight']=df['Weight'].fillna(method='bfill')
df['Number of days']=df['Number of days'].fillna(method='pad')
df['stay']=df['stay'].fillna(method='ffill') # chcek for bfill; in all the instances the value of stay will be 12.0
df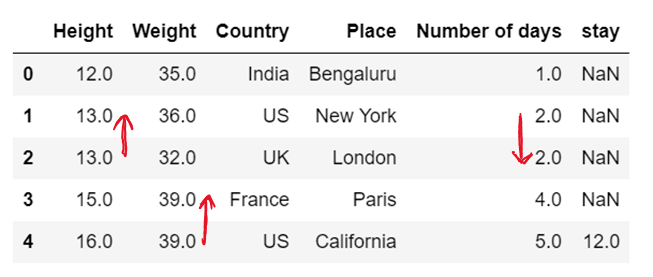
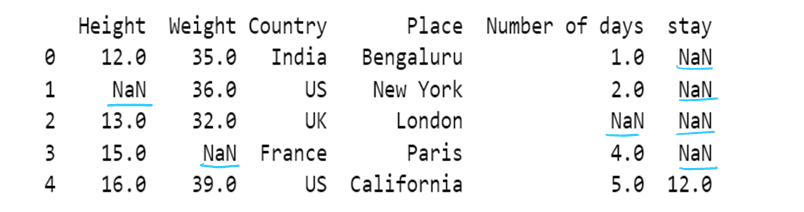
#c) using axis attribute
df3 = df3.fillna(method="backfill", axis=1)
print(df3)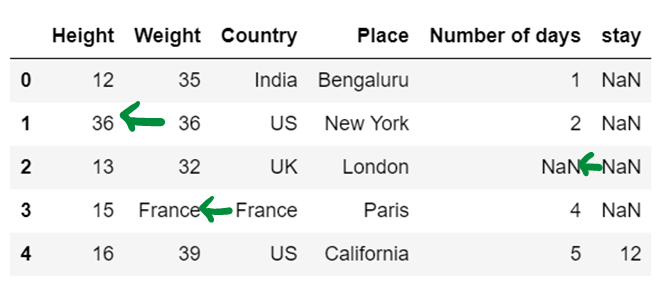
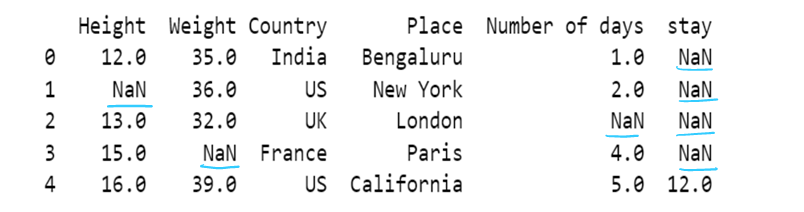
# Use of limit attribute ; if axis = 0--> in one column only one NaN value is replaced if limit = 1
if axis = 1--> in one row only one NaN value is replaced if limit = 1
df4 = df4.fillna(method="backfill", axis=0, limit = 1)
df4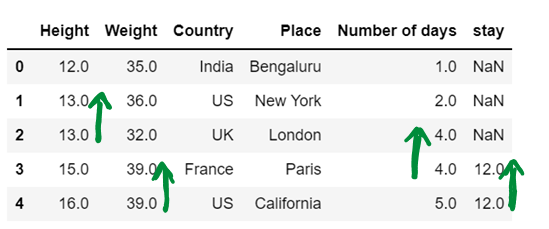
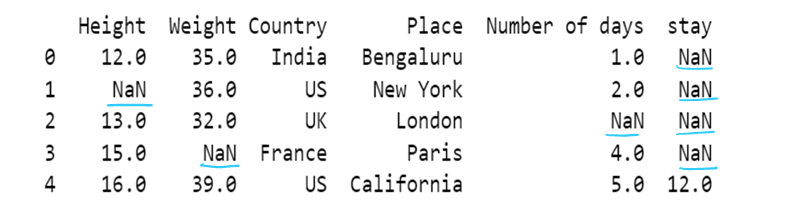
# Use of limit attribute ; if axis = 0--> in one column only one NaN value is replaced if limit = 1
if axis = 1--> in one row only one NaN value is replaced if limit = 1
df5 = df5.fillna(method="backfill", axis=1, limit = 1)
df5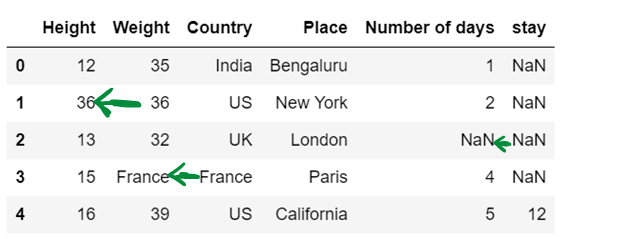
#f) Use of inplace. By default inplace = false. If inplace=true, then original dataframe is changed. else it remains unchanged
df6= pd.read_csv("F:/SRIHER/2021-2022/Quarter - 3/Advacned Python/Module - 1/Dataset/d1.csv")
print (df6)
df6.fillna(method="backfill",inplace = True)
df6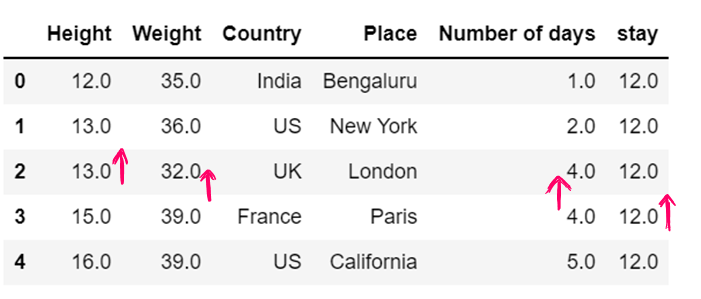
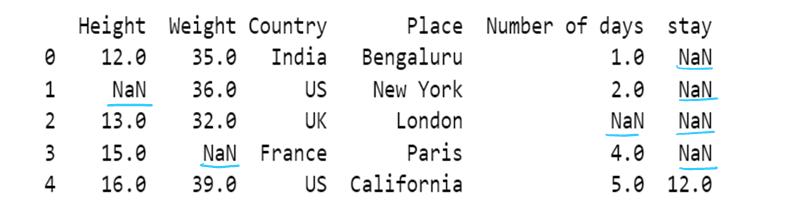
#f) Use of inplace. By default inplace = false. If inplace=true, then original dataframe is changed. else it remains unchanged
df6= pd.read_csv("F:/SRIHER/2021-2022/Quarter - 3/Advacned Python/Module - 1/Dataset/d1.csv")
print (df6)
df6.fillna(method="backfill",inplace = True)
df6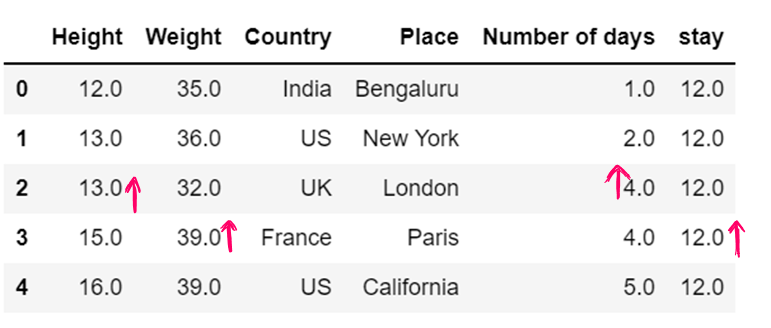
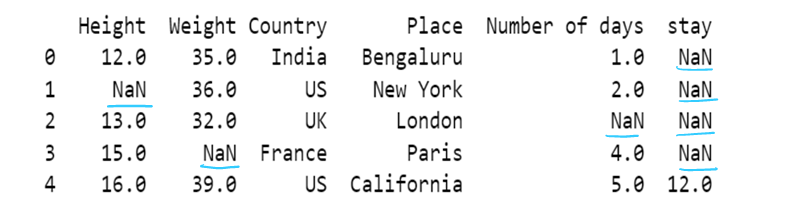
#g) Use of dictionary in value attribute
df7= pd.read_csv("F:/SRIHER/2021-2022/Quarter - 3/Advacned Python/Module - 1/Dataset/d1.csv")
print (df7)
values = {"Height": 0, "Weight": 1, "Country": 2, "Place": 3, "Number of days":4, "stay":5}
df7.fillna(value=values,inplace=True)
df7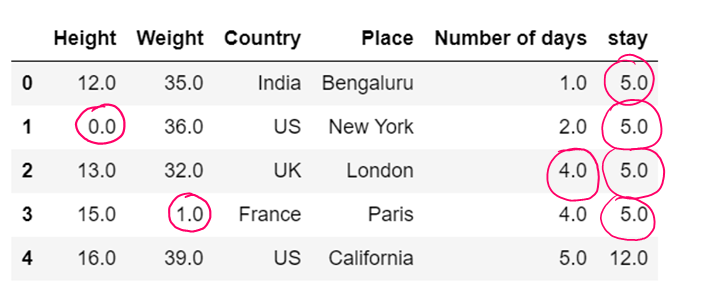
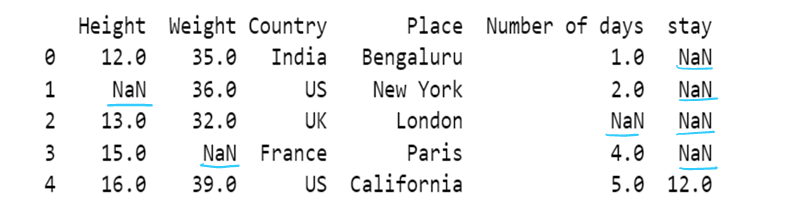
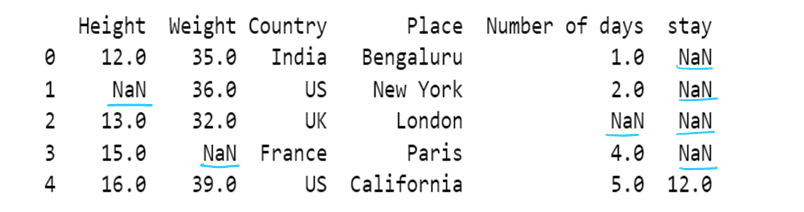
#g) Use of dictionary in value attribute with limit attribute
df8= pd.read_csv("F:/SRIHER/2021-2022/Quarter - 3/Advacned Python/Module - 1/Dataset/d1.csv")
print (df8)
values = {"Height": 0, "Weight": 1, "Country": 2, "Place": 3, "Number of days":4, "stay":5}
df8.fillna(value=values,inplace=True, limit=1)
df8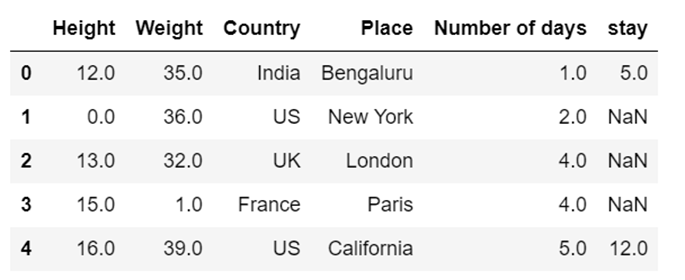
h) use of mean to impute the missing values
Fill NaN values in one column with mean:
df['col1'] = df['col1'].fillna(df['col1'].mean())
Fill NaN values in more than one column with mean:
df[['col1', 'col2']] = df[['col1', 'col2']].fillna(df[['col1', 'col2']].mean())
Fill NaN values in all column with mean:
df = df.fillna(df.mean())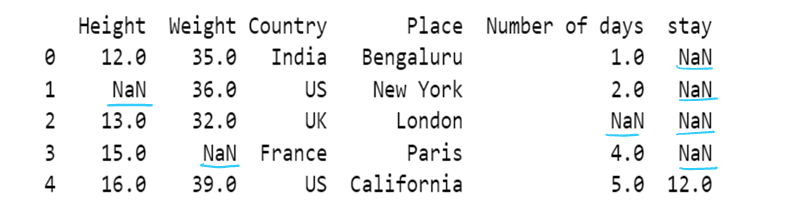
h) use of median to impute the missing values
Fill NaN values in one column with median:
df['col1'] = df['col1'].fillna(df['col1'].median())
Fill NaN values in more than one column with median:
df[['col1', 'col2']] = df[['col1', 'col2']].fillna(df[['col1', 'col2']].median())
Fill NaN values in all column with median:
df = df.fillna(df.median())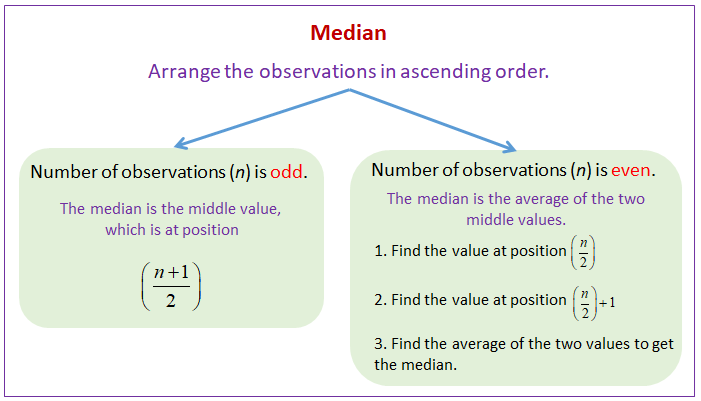
g) use of median to impute the missing values
df10['Weight']=df10['Weight'].fillna(df10['Weight'].median())
df10Average number is a-0 32 35 36 39 n = 4 Divide by 2 4/2 = 2 Get the number at index 2 i.e, 35
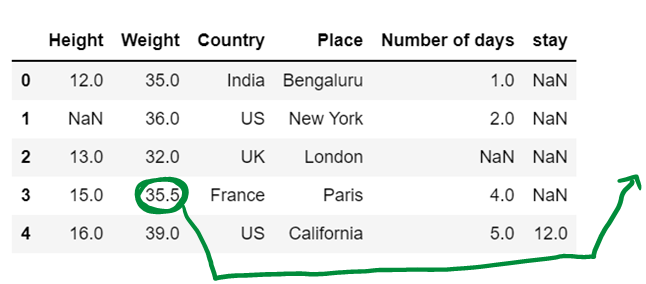
Get the next number and compute average (35+36) / 2
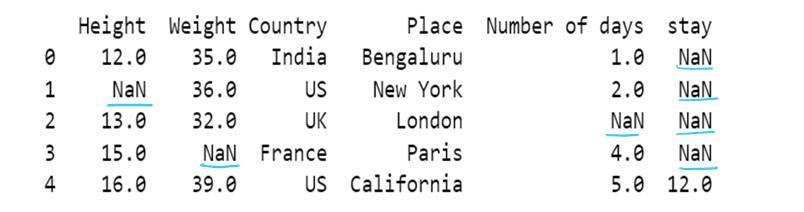
g) use of median to impute the missing values
df11[['Height','Weight']]=df11[['Height','Weight']].fillna(df11[['Height','Weight']].median())
df11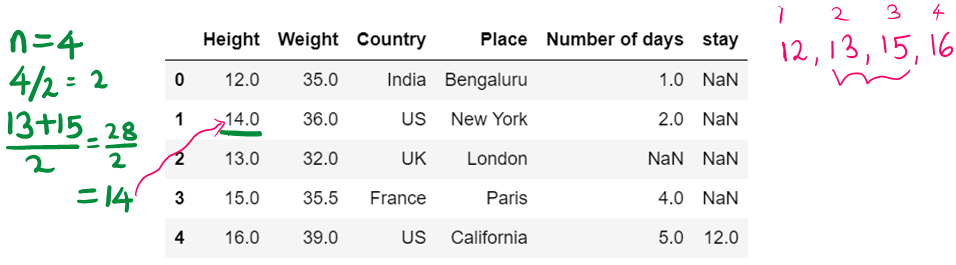
g) use of median to impute the missing values
df12=df12.fillna(df12.median())
df12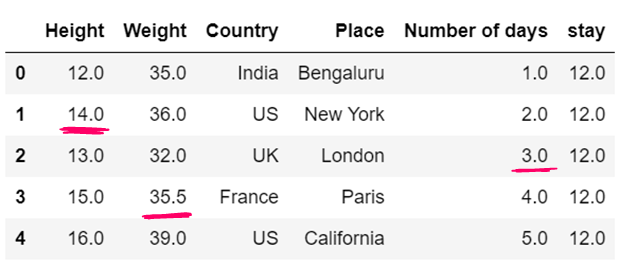
1, 2, 3, 4 n=4 4/2 = 2 Element at 2nd loc is 2 Next element is 4 = (2+4) = 6/2 = 3
- Mode is the most frequent observation (or observations) in a sample.
- EX: [5,2,3,3,4,6] – Mode is 3, because 3 appears two times in the sample whereas the other elements only appear once
- The mode does not have to be unique.
- Some samples have more than one mode
- Example: [5,2,3,3,4,6,5] Mode is 3 and 5 because they both appears more often and both appear same number of times
- The mode is commonly used for categorical data
- Boolean – take two values: true or false, male or female
- Nominal – Take more than two values : American, African, Asian
- Ordinal – Take more than two values but the values have a logical order: few- some – many
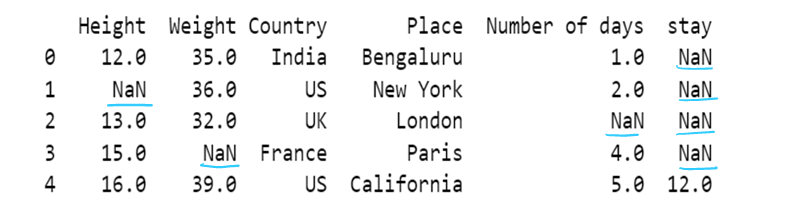
i) use of mode to impute the missing values
df13=df13.fillna(df13.mode())
df13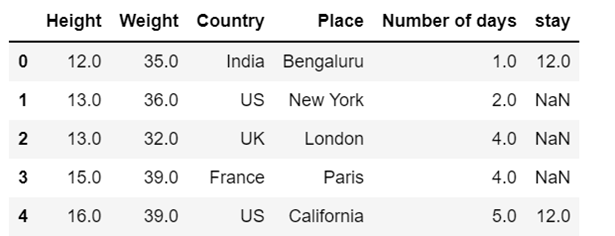
Views: 0